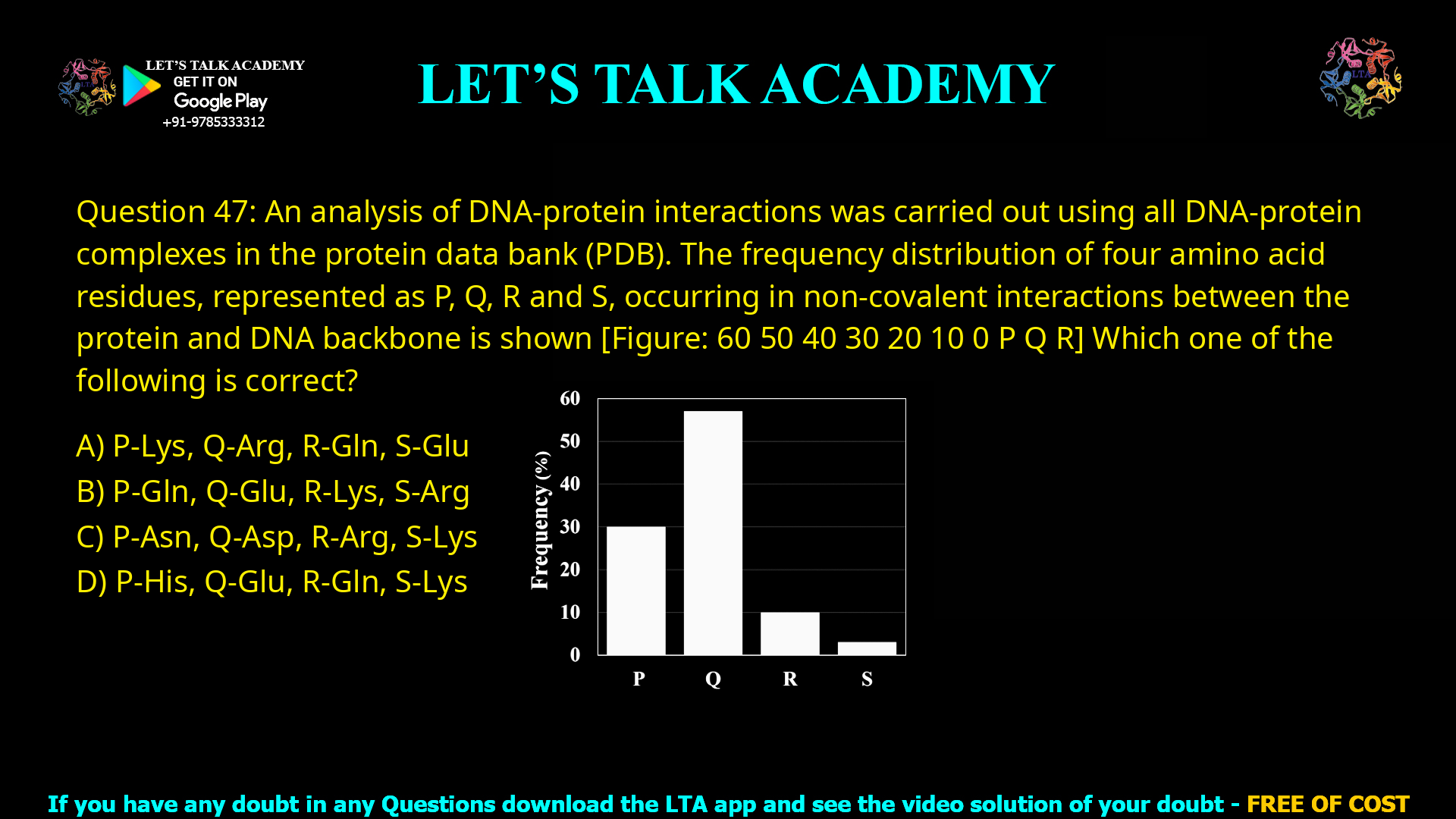
Q.47 An analysis of DNA-protein interactions was carried out using all DNA-protein complexes
in the protein data bank (PDB). The frequency distribution of four amino acid residues,
represented as P, Q, R and S, occurring in non-covalent interactions between the protein and
DNA backbone is shown below.
Which one of the following is correct?
(A) P-Lys, Q-Arg, R-Gln, S-Glu (B) P-Gln, Q-Glu, R-Lys, S-Arg
(C) P-Asn, Q-Asp, R-Arg, S-Lys (D) P-His, Q-Glu, R-Gln, S-Lys
Question Overview
A study of all DNA–protein complexes in the Protein Data Bank (PDB) analyzed which amino acids make non-covalent interactions with the DNA backbone.
A bar graph shows the frequency of four residues:
-
P ≈ 30%
-
Q ≈ 55%
-
R ≈ 10%
-
S ≈ 5%
We must match P, Q, R, S with the correct amino acids.
Concept Reminder
DNA backbone is negatively charged due to phosphate groups.
Therefore, amino acids that commonly bind DNA are:
-
Positively charged residues
-
Lysine (Lys, K)
-
Arginine (Arg, R)
-
-
Polar but uncharged residues
-
Glutamine (Gln, Q)
-
Asparagine (Asn, N)
-
Least frequent:
-
Acidic or negative residues (Asp, Glu)
Correct Answer: (A)
P–Lys, Q–Arg, R–Gln, S–Glu
Why Option (A) is Correct
Q has the highest frequency (~55%) → Arginine
Arginine interacts strongly with DNA due to multiple positive charges, enabling stable ionic interactions.
P is second (~30%) → Lysine
Another positively charged residue, Lysine, commonly binds to the negatively charged DNA phosphate backbone.
R is moderate (~10%) → Glutamine
Glutamine participates in hydrogen bonding, but less frequently than Lys/Arg.
S is minimal (~5%) → Glutamate
Glutamate is negatively charged, causing repulsion with DNA, explaining its low frequency.
Why Other Options Are Incorrect
(B) P–Gln, Q–Glu, R–Lys, S–Arg
-
Q cannot be Glu because Glutamate rarely binds DNA
-
S cannot be Arg because Arginine is the most frequent
(C) P–Asn, Q–Asp, R–Arg, S–Lys
-
Q as Asp is incorrect: Asp (negative) rarely binds DNA
-
R cannot be Arg because arginine should be highest
(D) P–His, Q–Glu, R–Gln, S–Lys
-
Glu again incorrectly placed at highest frequency
-
Lysine can’t be lowest (S)
Conclusion
The pattern of interaction frequency strongly favors positively charged residues, followed by polar, with acidic residues lowest.
Final Correct Answer: (A)
P–Lys, Q–Arg, R–Gln, S–Glu