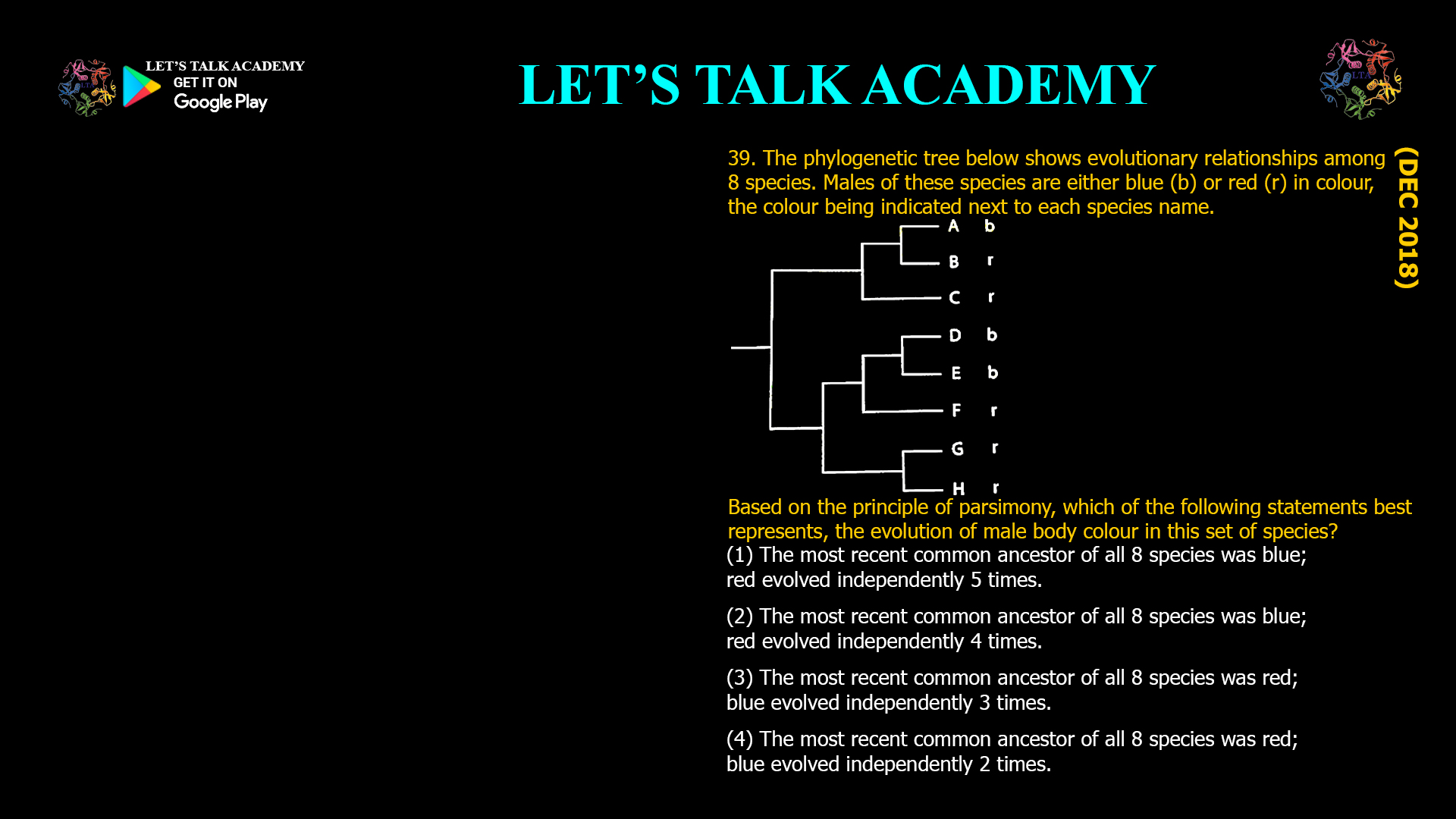
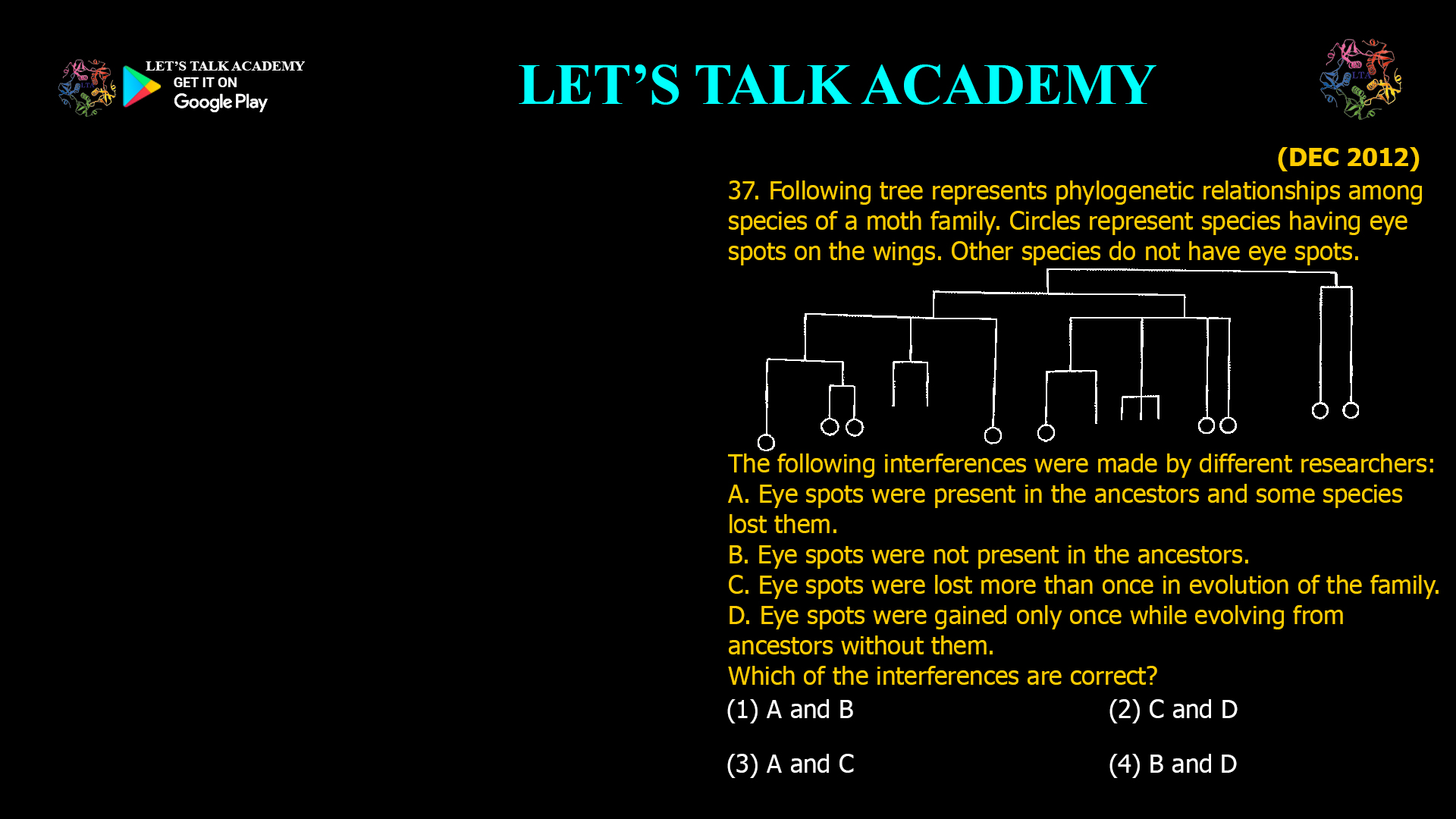
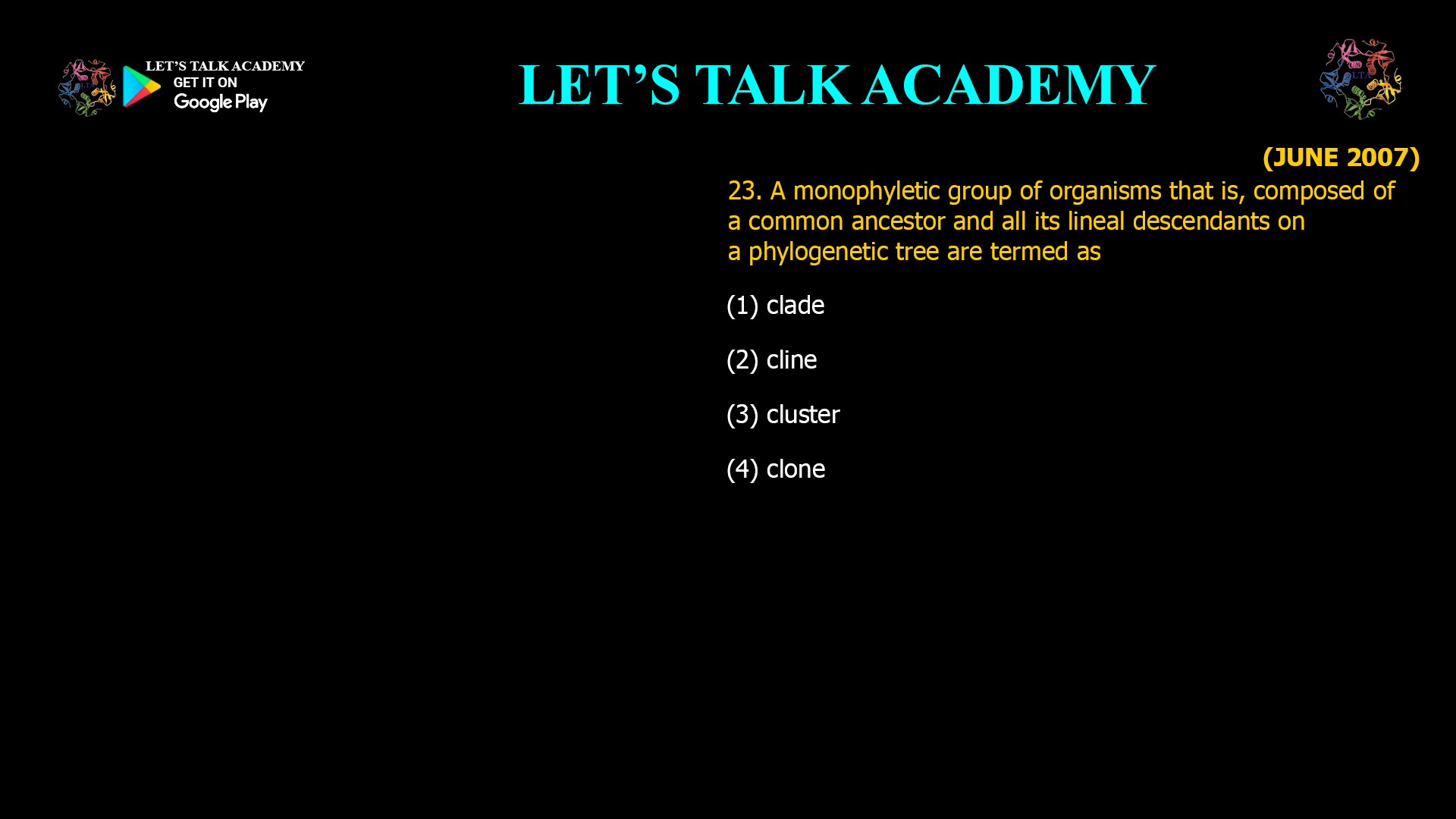
The two phylogenetic trees given below represent evolutionary patterns in species or population. The differently colored or dashed lines represent a single or gene genealogy. Select the option that correctly […]
Category: CSIR NET Life Science Previous Year Questions and Solution on Molecular Evolution
CSIR NET Life Science Previous Year Questions and Solution on Molecular Evolution