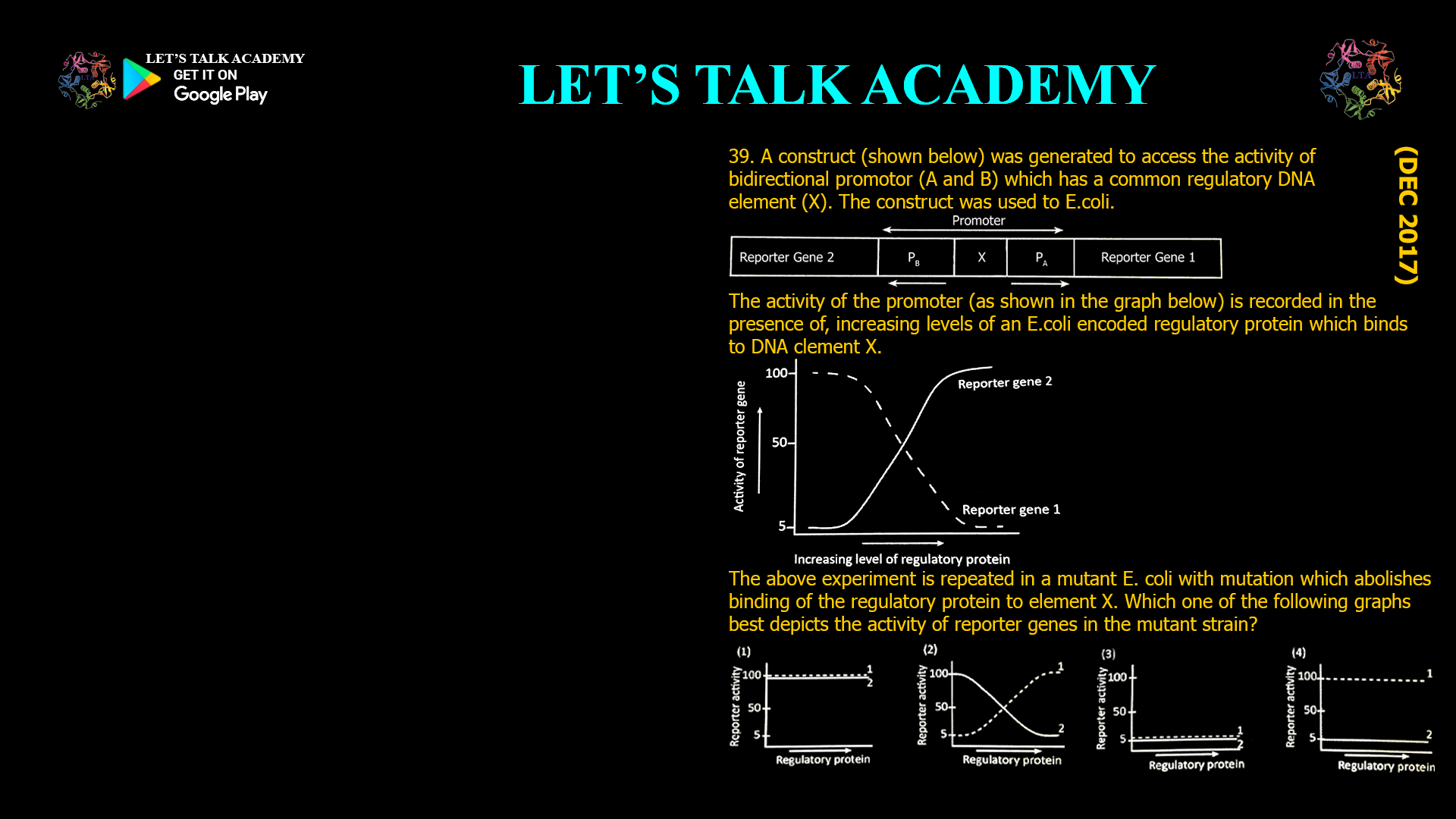
39. A construct (shown below) was generated
to access the activity of bidirectional
promotor (A and B) which has a common
regulatory DNA element (X). The construct
was used to E.coli.
The activity of the promoter (as shown in the
graph below) is recorded in the presence of,
increasing levels of an E.coli encoded
regulatory protein which binds to DNA
clement X.
The above experiment is repeated in a
mutant E. coli with mutation which abolishes
binding of the regulatory protein to element X.
Which one of the following graphs best
depicts the activity of reporter genes in the mutant strain
Bidirectional promoters are fascinating regulatory elements in bacterial genomes, allowing coordinated expression of two divergent genes from a single DNA region. In Escherichia coli, such promoters are not only common but also highly regulated, often sharing control elements that respond to specific regulatory proteins165. This article delves into the experimental analysis of a bidirectional promoter construct, the role of a common regulatory DNA element, and the impact of a mutation that abolishes regulatory protein binding on reporter gene activity.
Background: Bidirectional Promoters and Regulatory Elements
A bidirectional promoter is a DNA sequence that directs transcription in two opposite directions, enabling the simultaneous expression of two genes or reporter constructs. In E. coli, these promoters are often found between divergent genes and may share regulatory elements that respond to environmental signals or cellular conditions542. The activity of each promoter can be modulated by the binding of regulatory proteins to these shared elements, influencing the overall output of the reporter genes.
A typical experimental setup involves cloning a bidirectional promoter region—flanked by two reporter genes—into a plasmid and transforming it into E. coli. The activity of each reporter gene is then measured in response to varying concentrations of a regulatory protein that binds to a common DNA element (X) located between the two promoters. This approach allows researchers to dissect the regulatory logic of complex promoter architectures.
Experimental Setup: Measuring Promoter Activity
In the described experiment, a construct was generated containing a bidirectional promoter (A and B) with a common regulatory DNA element (X). The construct was introduced into E. coli, and the activity of each promoter was monitored using reporter genes (e.g., fluorescent proteins or enzymes whose activity can be quantified). The activity was measured as the concentration of an E. coli-encoded regulatory protein, which binds to element X, was increased.
In the wild-type scenario, the regulatory protein’s binding to element X alters the activity of both promoters A and B. Depending on whether the protein is an activator or a repressor, the reporter gene activity will increase or decrease as the protein concentration rises. This results in a characteristic curve on a graph, where the x-axis represents the increasing concentration of the regulatory protein, and the y-axis represents reporter gene activity.
Impact of Mutation: Loss of Regulatory Protein Binding
The experiment is then repeated in a mutant E. coli strain where the regulatory protein can no longer bind to element X. This mutation could be in the regulatory protein itself or in the DNA sequence of element X, but the outcome is the same: the regulatory protein is unable to exert its effect on the promoters.
In this mutant strain, the regulatory protein’s concentration no longer influences the activity of the reporter genes. As a result, the activity of both reporter genes remains constant, regardless of how much regulatory protein is present. This is because the critical link between the regulatory protein and the control of promoter activity has been broken.
Graphical Interpretation
The expected results can be summarized as follows:
-
Wild-type: The graph shows reporter gene activity changing with increasing regulatory protein concentration. If the regulatory protein is an activator, activity increases; if it is a repressor, activity decreases.
-
Mutant (no binding to X): The graph shows a flat line, indicating that reporter gene activity is unchanged as the regulatory protein concentration increases. The promoters operate at their basal levels, unaffected by the regulatory protein.
Biological Significance
The ability of regulatory proteins to modulate bidirectional promoter activity is crucial for fine-tuning gene expression in response to environmental cues. In natural systems, such regulation allows bacteria to adapt quickly to changes in nutrient availability, stress, or other signals165. Mutations that disrupt this regulation can lead to dysregulated gene expression, potentially affecting cellular fitness or metabolic efficiency.
Practical Applications
Understanding the regulation of bidirectional promoters is important for synthetic biology and metabolic engineering. By designing constructs with specific regulatory elements, researchers can create genetic circuits that respond predictably to external signals. Mutations that abolish regulatory protein binding can be used to study the contribution of specific regulatory interactions or to engineer strains with constitutive gene expression32.
Detailed Mechanism
The regulatory architecture of bidirectional promoters often involves overlapping or closely spaced promoter elements, with shared regulatory sequences. The binding of a regulatory protein to a common element can influence the recruitment or exclusion of RNA polymerase at both promoters, leading to coordinated changes in transcription167. The strength and position of the regulatory element relative to the promoters are critical for determining the magnitude and direction of the regulatory effect.
In the case of the fepD-ybdA bidirectional promoter in E. coli, for example, the binding of the Fur repressor to overlapping operator sequences modulates the activity of both promoters in response to iron availability16. Mutations that disrupt Fur binding abolish iron-mediated regulation, resulting in constitutive promoter activity.
Evolutionary Perspective
Bidirectional promoters are evolutionarily ancient and are thought to represent an early step in the evolution of gene regulation. Their prevalence in bacterial genomes, including E. coli, suggests that they provide a flexible and efficient means of coordinating gene expression54. The ability of regulatory proteins to modulate bidirectional promoter activity is likely under strong selective pressure, as it allows for rapid adaptation to changing environments.
Conclusion
In summary, when a mutation abolishes the binding of a regulatory protein to the common DNA element (X) in a bidirectional promoter construct, the activity of the reporter genes becomes independent of the regulatory protein concentration. The corresponding graph will show a flat line, indicating constant reporter gene activity regardless of the regulatory protein level. This contrasts with the wild-type, where reporter gene activity varies with regulatory protein concentration.
Key Takeaway:
In the mutant E. coli strain where the regulatory protein cannot bind to the shared DNA element, the activity of the reporter genes driven by the bidirectional promoter remains constant and is not influenced by the regulatory protein. The correct graph for this scenario is one where reporter gene activity does not change with increasing regulatory protein concentration.
This article explains how mutations affecting regulatory protein binding to a common DNA element alter bidirectional promoter activity and reporter gene expression in E. coli, providing clear guidance for interpreting experimental results and graphs.