- A 6.4 Kb plasmid DNA has two restriction endonuclease sites, HindIII and EcoRI. Complete double digestion of the plasmid with both the enzymes yields two fragments of 3.1 and 3.3 Kb. In order to study DNA repair process, a G:T m
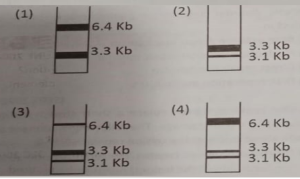
ismatch was introduced in one strand of HindIII site and the damaged plasmid was incubated in a reconstituted repair system containing all the factors and enzymes rquired for repair. If the efficiency of the repair system is 50%, which one of the following band patterns on agarose gel will be obtained after treating the repaired
DNA Repair Study Usin
Studying DNA repair mechanisms often involves introducing specific lesions into plasmid DNA and analyzing repair efficiency by restriction enzyme digestion and gel electrophoresis. This article discusses a scenario where a 6.4 Kb plasmid with two restriction sites—HindIII and EcoRI—is modified by introducing a G:T mismatch at the HindIII site. After incubation in a repair system with 50% efficiency, the resulting digestion pattern on an agarose gel is predicted.
Background: Plasmid and Restriction Sites
-
Plasmid size: 6.4 Kb
-
Restriction sites: HindIII and EcoRI, yi
-
3.1 Kb fragment
-
3.3 Kb fragment
-
-
elding two fragments upon double digestion:
Effect of G:T Mismatch at HindIII Site
-
The HindIII recognition sequence is palindromic (5′-AAGCTT-3′).
-
A G:T mismatch in one strand
-
of the HindIII site disrupts the recognition sequence.
-
Restriction enzymes like HindIII are highly sequence-specific and generally do not cut if their recognition site is mutated or mismatched.
-
EcoRI site remains intact and cuts normally.
Repair and Digestion Outcomes
-
The plasmid is incubated in a repair system with 50% efficiency to correct the G:T mismatch back to the correct G:C base pair, restoring the HindIII site in half of the plasmid molecules.
-
After repair, the sample is digested with both HindIII and EcoRI.
Expected Band Patterns on Agarose Gel
-
Unrepaired plasmids (50%):
-
HindIII site remains mutated (no cleavage at HindIII site).
-
EcoRI site is cleaved normally.
-
Result: One linear fr
-
agment of approximately 6.4 Kb (since only EcoRI cuts once, the plasmid remains circular or nicked but not cut into two fragments).
-
-
Repaired plasmids (50%):
-
HindIII site restored and cleaved normally.
-
EcoRI site cleaved normally.
-
Result: Two fragments of 3.1 Kb and 3.3 Kb.
-
Overall Gel Pattern
-
Three bands expected:
-
One band at ~6.4 Kb (uncut or single-cut plasmid) from unrepaired plasmids.
-
Two bands at 3.1 Kb and 3.3 Kb fro
-
m completely digested, repaired plasmids.
-
-
The intensity of the 6.4 Kb band and the sum of the 3.1 Kb and 3.3 Kb bands should be approximately equal, reflecting 50% repair efficiency.
Summary Table of Bands
Plasmid Population HindIII Site Status Digestion Pattern Band Sizes on Gel 50% unrepaired Mutated (no cut) Only EcoRI cuts Single ~6.4 Kb band 50% repaired Restored (cut) Both HindIII and EcoRI cut Two bands: 3.1 Kb and 3.3 Kb
Keywords for SEO Optimization
-
Plasmid DNA repair assay
-
HindIII and EcoRI digestion
-
G:T mismatch repair
-
DNA repair efficiency assay
-
Restriction enzyme site mutation
-
Agarose gel electrophoresis DNA fragments
-
DNA mismatch repair system
-
DNA repair and restriction digestion
-
Molecular cloning and DNA repair
-
DNA lesion correction assay
Conclusion
When a 6.4 Kb plasmid containing HindIII and EcoRI sites is damaged by a G:T mismatch at the HindIII site and repaired with 50% efficiency, digestion with both enzymes yields a gel pattern with:
-
A 6.4 Kb band from unrepaired plasmids (HindIII site mutated, no cut).
-
Two bands of 3.1 Kb and 3.3 Kb from repaired plasmids (both sites cut).
This pattern reflects the partia
-



4 Comments
Komal Sharma
August 22, 2025Option 3 is correct
Ajay Sharma
September 4, 2025Good explanation
Neelam Sharma
November 8, 2025Option 3 is correct
Parul
November 11, 2025Option – 4 is correct
Three bands – 6.4Kb, 3.1Kb and 3.3Kb.
The intensity of the 6.4Kb band and the sum of the 3.1Kb and 3.3Kb bands should be approximately equal.