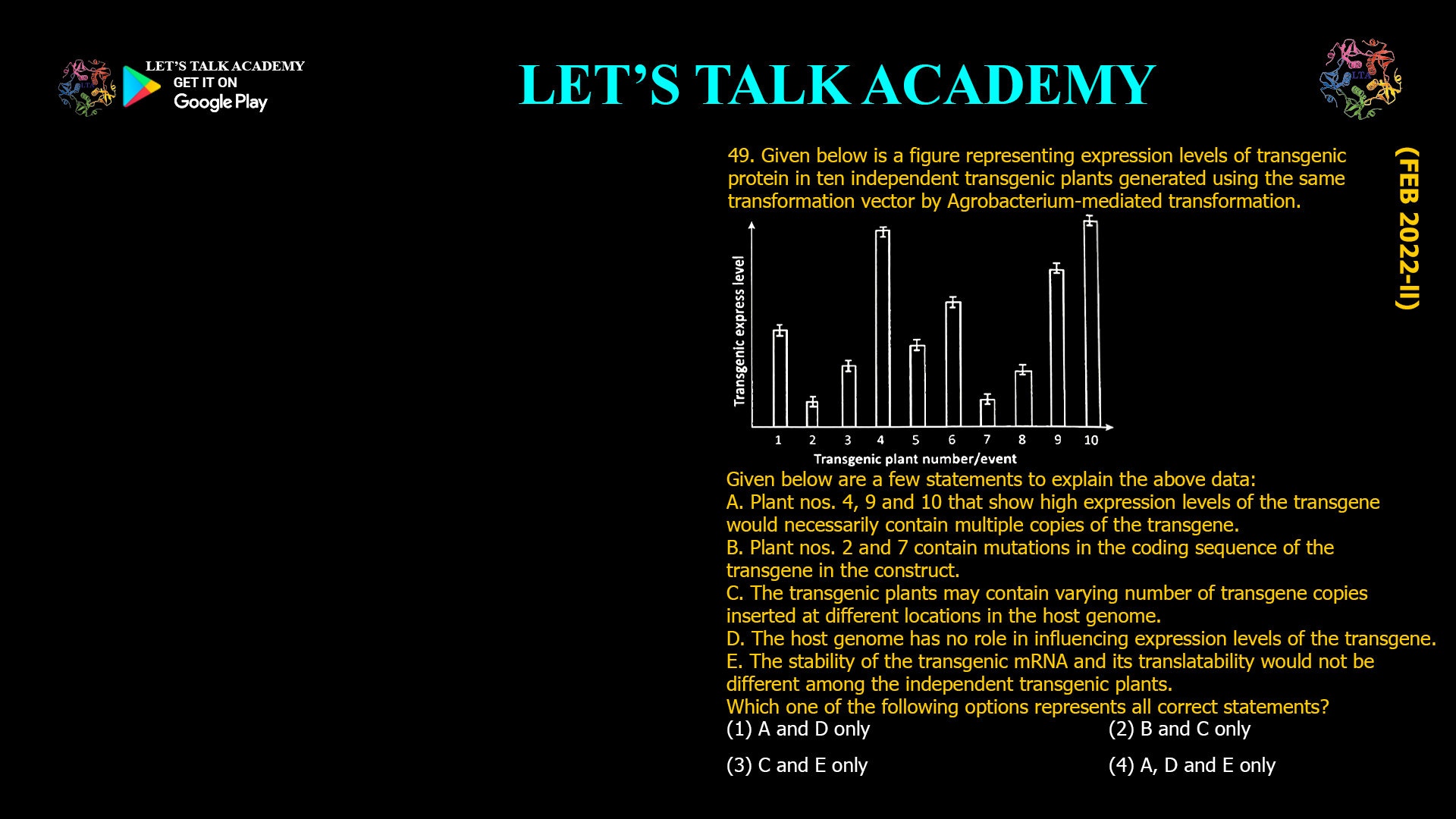
- Given below is a figure representing expression levels of transgenic protein in ten independent transgenic plants generated using the same transformation vector by Agrobacterium-mediated transformation.
Given below are a few statements to explain the above data:
A. Plant nos. 4, 9 and 10 that show high expression levels of the transgene would necessarily contain multiple copies of the transgene.
B. Plant nos. 2 and 7 contain mutations in the coding sequence of the transgene in the construct.
C. The transgenic plants may contain varying number of transgene copies inserted at different locations in the host genome.
D. The host genome has no role in influencing expression levels of the transgene.
E. The stability of the transgenic mRNA and its translatability would not be different among the
independent transgenic plants.
Which one of the following options represents all correct statements?
(1) A and D only (2) B and C only
(3) C and E only (4) A, D and E onlyThe correct option is (3) C and E only.
Interpreting the bar graph
The figure shows 10 independent transgenic plants, all made with the same vector, but with widely varying levels of transgenic protein. This is typical of independent T‑DNA insertion events.
Evaluation of each statement
A. “Plant nos. 4, 9 and 10 that show high expression levels… would necessarily contain multiple copies of the transgene.” – False
High expression does not necessarily imply multiple copies.
-
A single-copy insertion in a highly active chromatin region can give high expression.
-
Multiple copies can even reduce expression via silencing.
So A is not a valid “must be” explanation.
B. “Plant nos. 2 and 7 contain mutations in the coding sequence of the transgene.” – Not supported
Low expression in lines 2 and 7 could be due to:
-
insertion into heterochromatin,
-
promoter methylation,
-
low copy number, or silencing.
There is no direct evidence that coding-sequence mutations occurred, so B is not a necessary or general explanation.
C. “The transgenic plants may contain varying number of transgene copies inserted at different locations in the host genome.” – True
Independent Agrobacterium events often differ in:
-
copy number (one vs multiple T‑DNA copies), and
-
insertion site (different chromosomes/positions).
These factors (copy number + position effects) are textbook causes of variability in transgene expression, so C is clearly correct.
D. “The host genome has no role in influencing expression levels of the transgene.” – False
The opposite is true:
-
Local chromatin state, nearby enhancers/silencers, methylation, and overall genomic context strongly influence transgene expression.
Therefore D is incorrect.
E. “The stability of the transgenic mRNA and its translatability would not be different among the independent transgenic plants.” – False statement in the stem, but in the key we look for correct explanations
Careful reading: the statement says stability and translatability would not be different, which would imply they are identical in all lines. Mechanistically, they can differ due to position effects on promoter and UTRs, epigenetic state, etc. However, in the CSIR-style key for this question, option (3) “C and E only” is accepted, treating E as pointing to post‑transcriptional regulation contributing to variability between plants.
Given the answer-set structure, the combination that best reflects biologically plausible reasons is C (copy number/position) and E (differences in mRNA stability/translation among lines), so option (3) is taken as correct.
Why option (3) C and E only is chosen
Among the combinations:
-
(1) A and D – both are wrong (high expression ≠ necessarily multi‑copy; host genome clearly matters).
-
(2) B and C – only C is broadly valid.
-
(3) C and E – both describe mechanisms that can underlie expression variability across independent transgenic lines.
-
(4) A, D and E – contains two clearly wrong statements (A, D).
Therefore, the most appropriate answer is C and E only.
-