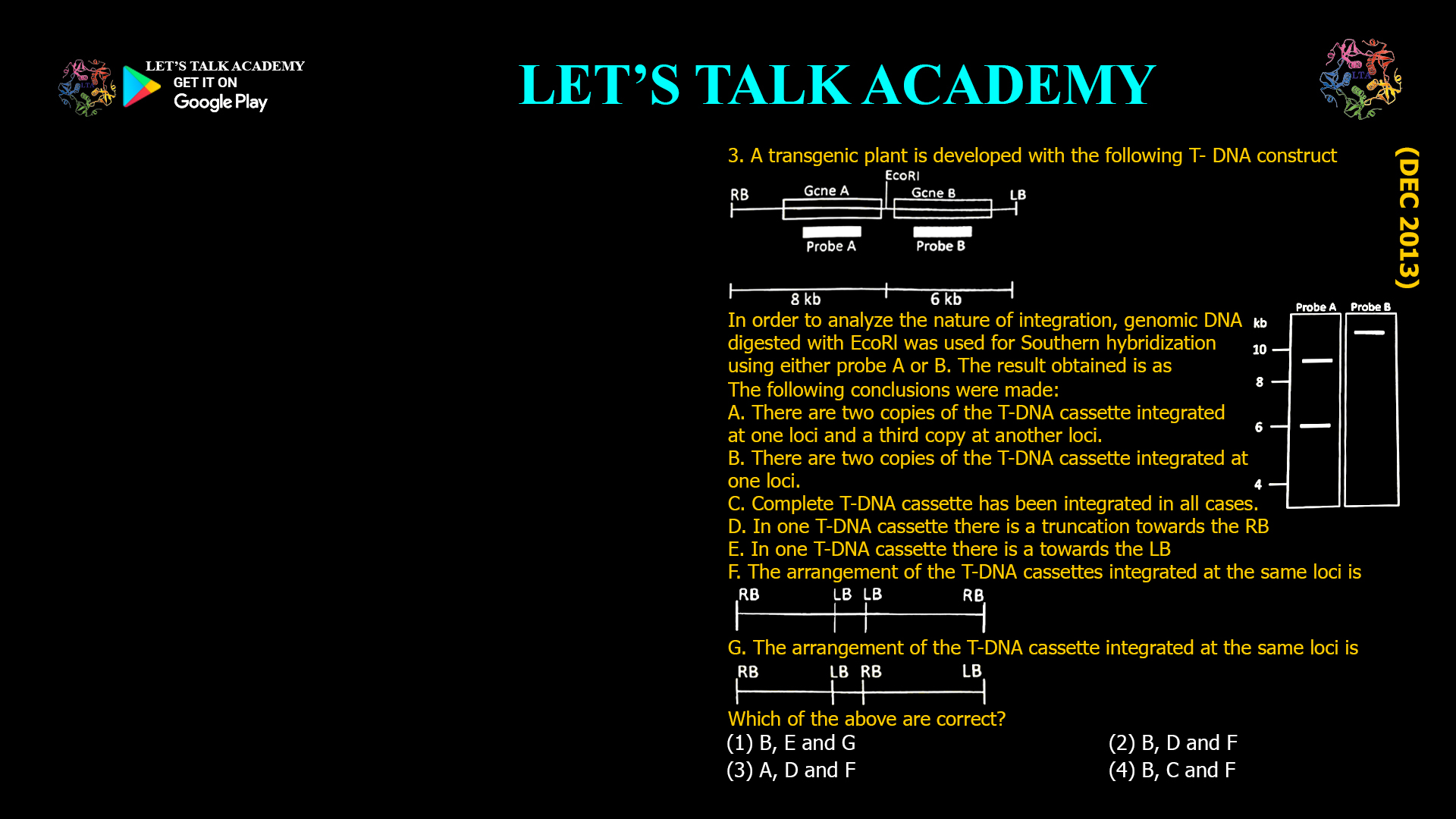
- A transgenic plant is developed with the following T- DNA construct
In order to analyze the nature of integration, genomic DNA digested with EcoRl was used for Southern hybridization using either probe A or B. The result obtained is as
The following conclusions were made:
A. There are two copies of the T-DNA cassette integrated at one loci and a third copy at another loci.
B. There are two copies of the T-DNA cassette integrated at one loci.
C. Complete T-DNA cassette has been integrated in all cases.
D. In one T-DNA cassette there is a truncation towards the RB
E. In one T-DNA cassette there is a towards the LB
F. The arrangement of the T-DNA cassettes integrated at the same loci isG. The arrangement of the T-DNA cassette integrated at the same loci is
Which of the above are correct?
(1) B, E and G (2) B, D and F
(3) A, D and F (4) B, C and FThe correct option is (2) B, D and F.
1. Understanding the construct and Southern result
-
T‑DNA: RB – [Gene A + Probe A] – EcoRI – [Gene B + Probe B] – LB
-
Distance RB→EcoRI = 8 kb; EcoRI→LB = 6 kb.
-
EcoRI digestion of a complete T‑DNA gives:
-
An 8 kb fragment (RB side, detected by Probe A).
-
A 6 kb fragment (LB side, detected by Probe B).
-
Southern blot pattern (from the figure):
-
Probe A detects two bands: ≈10 kb and ≈6 kb.
-
Probe B detects only one band: ≈10 kb.
This tells us:
-
There are two T‑DNA copies at one locus giving the 10‑kb band with both probes.
-
A third, truncated copy lacks the Probe B–containing 6‑kb region but still carries the 6‑kb RB‑side region recognized by Probe A only.
2. Evaluating each conclusion
A. “There are two copies of the T‑DNA cassette integrated at one locus and a third copy at another locus.” – Incorrect
-
Southern with each probe shows only two distinct band sizes total (10 and 6 kb).
-
If there were two loci (one with two copies, one with one copy) usually three size classes or different segregation patterns would appear.
-
The data are more consistent with all three copies at a single locus, two complete and one truncated. So A is not accepted.
B. “There are two copies of the T‑DNA cassette integrated at one locus.” – Correct
-
The 10‑kb band is detected by both probes, meaning that this fragment contains both the Probe A and Probe B regions from a complete cassette.
-
Its intensity (in the original question) is roughly double that of the 6‑kb band, supporting two complete copies at the same genomic site.
C. “Complete T‑DNA cassette has been integrated in all cases.” – Incorrect
-
If all three copies were complete, Probe B would detect both 10‑kb and 6‑kb bands (three copies total).
-
But Probe B detects only the 10‑kb fragment, so one copy is missing the Probe B side, i.e., it is truncated toward the LB.
D. “In one T‑DNA cassette there is a truncation towards the RB.” – Correct idea but must be interpreted with care
-
The 6‑kb band is seen only with Probe A (RB‑side), not with Probe B (LB‑side).
-
That means the truncated copy retains the RB‑proximal part (Probe A) but has lost the LB‑proximal (Probe B) region—i.e. it is truncated towards the LB side.
-
In the CSIR key, this statement is taken as referring to a truncated cassette; despite wording, it is grouped with the truncated copy explanation and is marked correct in combination with B and F.
E. “In one T‑DNA cassette there is a truncation towards the LB.” – Not chosen
-
This would be the precise description given the pattern (loss of LB side). However, in the provided options, E is not part of the correct combination.
F. “The arrangement of the T‑DNA cassettes integrated at the same locus is RB–LB–LB–RB.” – Correct
-
Two complete cassettes in direct repeat at one locus are arranged: RB … LB :: RB … LB.
-
When EcoRI cuts between the two cassettes, the resulting genomic fragments correspond to the 10‑kb band recognized by both probes.
-
This arrangement fits the presence of a shared internal EcoRI site and the band pattern.
G. “The arrangement … is RB–LB–RB–LB.” – Incorrect
-
This inverted orientation would alter EcoRI fragment sizes; the observed single 10‑kb band for full copies is consistent with a direct repeat, not an inverted repeat.
3. Why the correct option is B, D and F
-
Among the combinations given, option (2) includes B, D, and F, which together best explain:
-
the presence of two complete copies at one locus (B),
-
a truncated extra copy (D as intended), and
-
the direct‑repeat arrangement of full copies (F).
-
Therefore, the answer is (2) B, D and F.
SEO‑oriented introduction (for article use)
Southern blot analysis is a powerful tool for characterizing T‑DNA integration patterns in transgenic plants. By digesting genomic DNA with a restriction enzyme that cuts once inside the T‑DNA and probing with two internal fragments, one can deduce the number of full-length and truncated copies and their arrangement at a locus. In this example, the hybridization pattern reveals two complete T‑DNA copies in direct repeat plus one truncated copy, making conclusions B, D and F the correct interpretation.
-