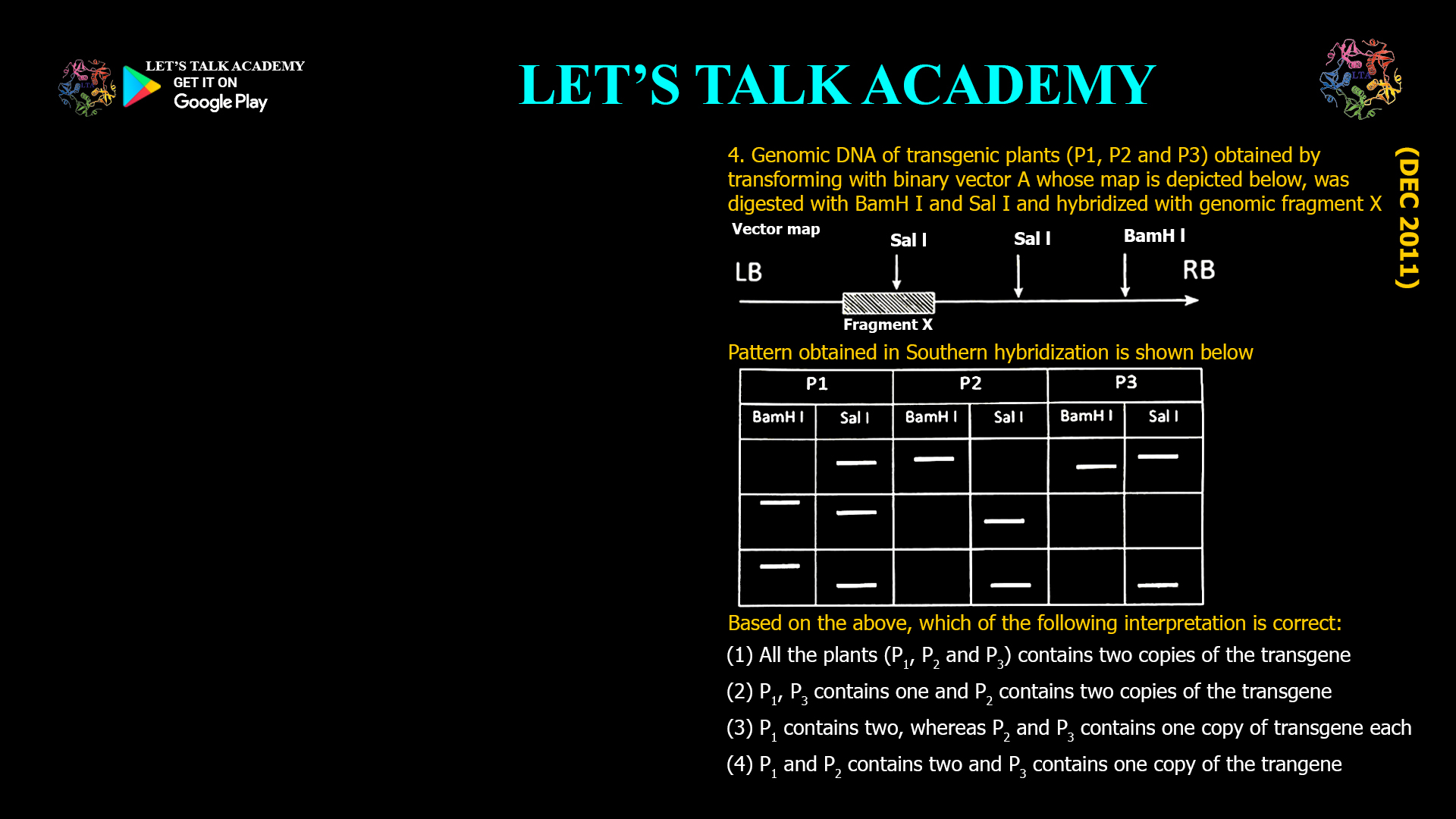
- Genomic DNA of transgenic plants (P1, P2 and P3) obtained by transforming with binary vector A whose map is depicted below, was digested with BamH I and Sal I and hybridized with genomic fragment X
Pattern obtained in Southern hybridization is shown below
Based on the above, which of the following interpretation is correct:
(1) All the plants (P1, P2 and P3) contains two copies of the transgene
(2) P1, P3 contains one and P2 contains two copies of the transgene
(3) P1 contains two, whereas P2 and P3 contains one copy of transgene each
(4) P1 and P2 contains two and P3 contains one copy of the trangeneThe correct interpretation is (3) P₁ contains two copies, whereas P₂ and P₃ contain one copy of the transgene each.
1. Understanding the setup
-
Binary vector map: LB –– [T‑DNA with internal fragment X] –– RB.
-
Within this T‑DNA there is one SalI site to the right of fragment X and a BamHI site further right (still inside T‑DNA).
-
Genomic DNA from three plants (P₁, P₂, P₃) is cut separately with BamHI or SalI and probed with fragment X (internal to the cassette).
-
Each independent insertion produces one fragment containing fragment X for each restriction enzyme digestion.
-
So, number of hybridizing bands with a given enzyme ≈ copy number of the transgene.
-
From the Southern table in the figure (qualitative):
-
P₁:
-
BamHI lane: two bands.
-
SalI lane: two bands.
-
-
P₂:
-
BamHI: one band.
-
SalI: one band.
-
-
P₃:
-
BamHI: one band.
-
SalI: one band.
-
Thus, P₁ carries two independent transgene copies; P₂ and P₃ each carry one.
2. Evaluating each option
(1) “All the plants (P₁, P₂, P₃) contain two copies of the transgene.” – Incorrect
-
P₂ and P₃ show only one band per enzyme, so they each have one copy, not two.
(2) “P₁, P₃ contain one and P₂ contains two copies.” – Incorrect
-
This reverses what the blot shows: the double‑band pattern is in P₁, not P₂ or P₃.
(3) “P₁ contains two, whereas P₂ and P₃ contain one copy each.” – Correct
-
Matches the observation:
-
Two bands (BamHI and SalI) in P₁ → two insertions.
-
Single band in P₂ and P₃ → single insertion in each.
-
(4) “P₁ and P₂ contain two and P₃ contains one copy.” – Incorrect
-
Again contradicts the Southern pattern; P₂ does not show two bands.
3. Key reasoning point
For an internal probe like fragment X, every independent T‑DNA insertion yields one BamHI fragment and one SalI fragment that hybridize. Counting the number of bands with either enzyme directly indicates the copy number. P₁ has two such fragments; P₂ and P₃ have one each, hence option (3).
SEO‑oriented introduction (for article use)
Southern blotting remains a standard way to estimate transgene copy number in transformed plants. When genomic DNA from P₁, P₂ and P₃ is digested with BamHI or SalI and probed with an internal T-DNA fragment X, the number of hybridizing bands reveals the number of independent insertions. The pattern—two bands in P₁ and single bands in P₂ and P₃—shows that P₁ carries two copies of the transgene, while P₂ and P₃ each carry one copy, making option (3) the correct interpretation.
-