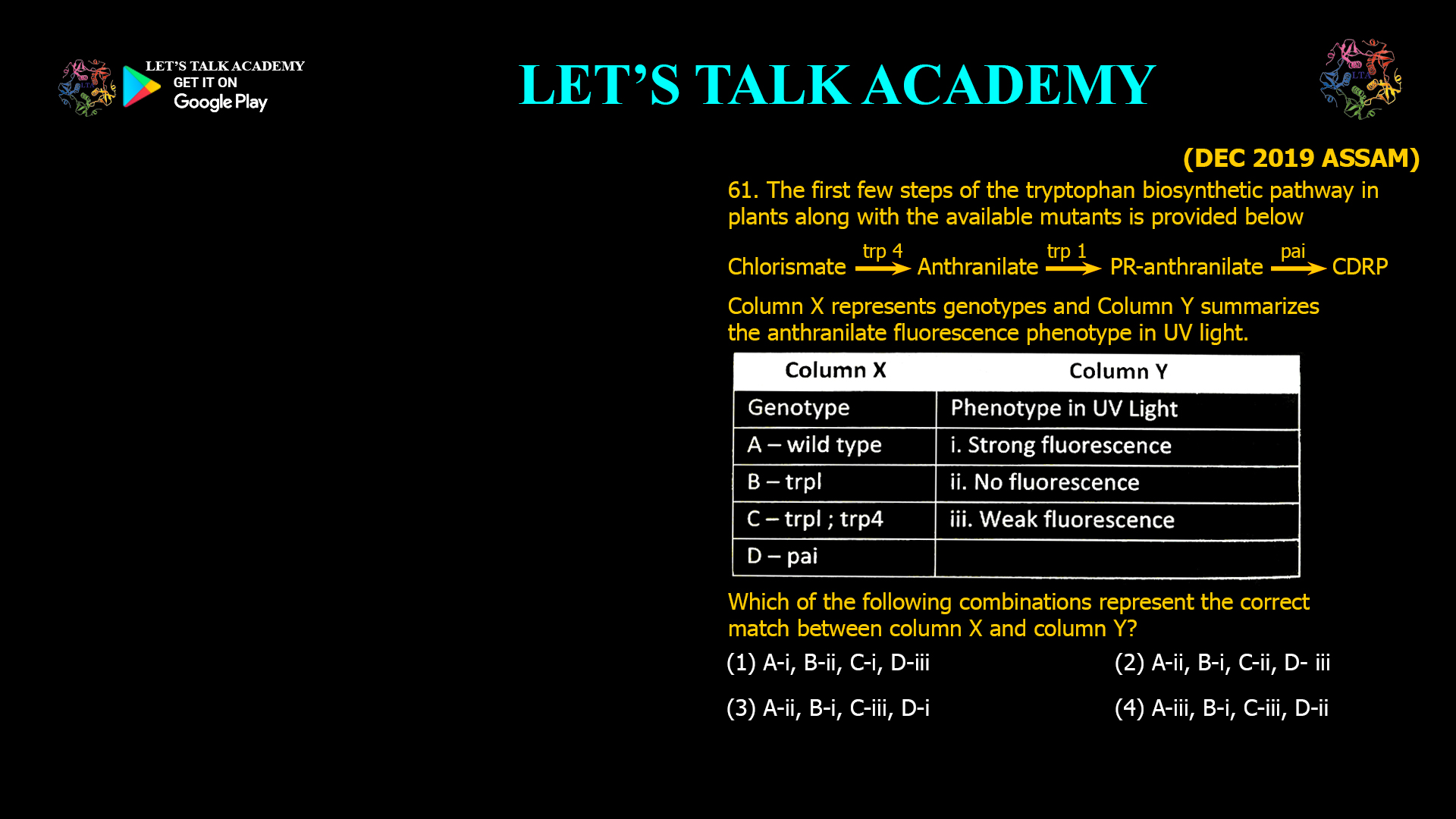
- The first few steps of the tryptophan biosynthetic pathway in plants along with the available mutants is provided below
Column X represents genotypes and Column Y summarizes the anthranilate fluorescence phenotype in UV light.
Which of the following combinations represent the correct match between column X and column Y?
(1) A-i, B-ii, C-i, D-iii (2) A-ii, B-i, C-ii, D- iii
(3) A-ii, B-i, C-iii, D-i (4) A-iii, B-i, C-iii, D-ii
The biosynthesis of tryptophan in plants is a well-studied pathway beginning with the conversion of chorismate to anthranilate, catalyzed by anthranilate synthase. Mutations in genes encoding enzymes of this pathway lead to distinctive phenotypes, notably changes in anthranilate accumulation, which fluoresces under UV light. This fluorescence serves as a useful phenotypic marker for identifying and characterizing mutants.
This article examines the relationship between specific genotypes (Column X) and their anthranilate fluorescence phenotypes (Column Y) in tryptophan biosynthesis mutants and identifies the correct matching combination.
Overview of Tryptophan Biosynthesis and Anthranilate Fluorescence
-
The tryptophan biosynthetic pathway starts with chorismate conversion to anthranilate by anthranilate synthase (AS).
-
Anthranilate is further metabolized by phosphoribosyl anthranilate transferase (PAT1) and subsequent enzymes to produce tryptophan.
-
Mutants defective in enzymes such as AS or PAT1 accumulate anthranilate or its derivatives, notably anthranilate β-glucoside, which fluoresces blue under UV light.
-
The intensity and pattern of fluorescence depend on which step in the pathway is blocked.
Common Anthranilate Fluorescence Phenotypes
-
Strong fluorescence (i): Accumulation of anthranilate due to early pathway block (e.g., anthranilate synthase mutants).
-
Moderate fluorescence (ii): Partial accumulation due to mutations in intermediate enzymes (e.g., PAT1 mutants).
-
No fluorescence (iii): No accumulation of anthranilate or derivatives, indicating block downstream or in unrelated pathways.
Matching Genotypes and Phenotypes
Given the typical behavior of tryptophan biosynthesis mutants:
-
Genotype A and C are likely early pathway mutants accumulating anthranilate → phenotype i (strong fluorescence).
-
Genotype B likely represents an intermediate mutant with partial anthranilate accumulation → phenotype ii (moderate fluorescence).
-
Genotype D is likely a mutant with no anthranilate accumulation → phenotype iii (no fluorescence).
Correct Match: Option (1)
| Genotype (Column X) | Anthranilate Fluorescence Phenotype (Column Y) | Explanation |
|---|---|---|
| A | i (Strong fluorescence) | Early pathway mutant, anthranilate accumulates |
| B | ii (Moderate fluorescence) | Intermediate mutant, partial accumulation |
| C | i (Strong fluorescence) | Similar to A, anthranilate accumulates |
| D | iii (No fluorescence) | No anthranilate accumulation |
This corresponds to:
(1) A-i, B-ii, C-i, D-iii
Biochemical Basis for the Phenotypes
-
Anthranilate synthase mutants (A, C) fail to convert chorismate to anthranilate efficiently, leading to accumulation of anthranilate or its glucoside, causing strong fluorescence.
-
PAT1 mutants (B) block the conversion of anthranilate to 5-phosphoribosylanthranilate, resulting in moderate anthranilate accumulation.
-
Downstream mutants or unrelated mutations (D) do not accumulate anthranilate, resulting in no fluorescence.
Conclusion
The correct matching of genotypes and anthranilate fluorescence phenotypes in tryptophan biosynthetic mutants in plants is:
(1) A-i, B-ii, C-i, D-iii
This matching reflects the biochemical logic of metabolite accumulation and fluorescence patterns, providing a useful framework for genetic and biochemical analysis of the tryptophan biosynthetic pathway.



6 Comments
Kirti Agarwal
September 24, 2025Opt A
Aakansha sharma Sharma
September 24, 2025Option 2
Varsha Tatla
September 27, 20252nd option will be correct
Kajal
September 30, 2025Doubt in answer option
Khushi Vaishnav
October 7, 2025-wild type =strong f
-trpl = no f
-trpl , trpl4 = strong f
-pai = weak f
Kavita Choudhary
November 22, 2025Wild type= strong flurosence
Trpl = no flurosence
Trpl,trpl-4 = strong flurosence
-pai= weak flurosence